Multiclass classification
Several SIMCA models can be combined to a special object simcam, which is used to make a multiclass classification. Besides this, it also allows calculating distance between individual models and a discrimination power — importance of variables to discriminate between any two classes. Let’s see how it works.
First we create three single-class SIMCA models with individual settings, such as number of optimal components and alpha.
m.set = simca(X.set, "setosa", 3, alpha = 0.01)
m.set = selectCompNum(m.set, 1)
m.vir = simca(X.vir, "virginica", 3)
m.vir = selectCompNum(m.vir, 2)
m.ver = simca(X.ver, "versicolor", 3)
m.ver = selectCompNum(m.ver, 1)Then we combine the models into a simcam model object. Summary will show the performance on
calibration set, which is a combination of calibration sets for each of the individual models
##
## SIMCA multiple classes classification (class simcam)
##
## Number of classes: 3
## Info:
##
## Summary for calibration results
## Ncomp TP FP TN FN Spec. Sens. Accuracy
## setosa 1 25 0 50 0 1.00 1.00 1.00
## virginica 2 22 3 47 3 0.94 0.88 0.92
## versicolor 1 25 3 47 0 0.94 1.00 0.96Now we apply the combined model to the test set and look at the predictions.
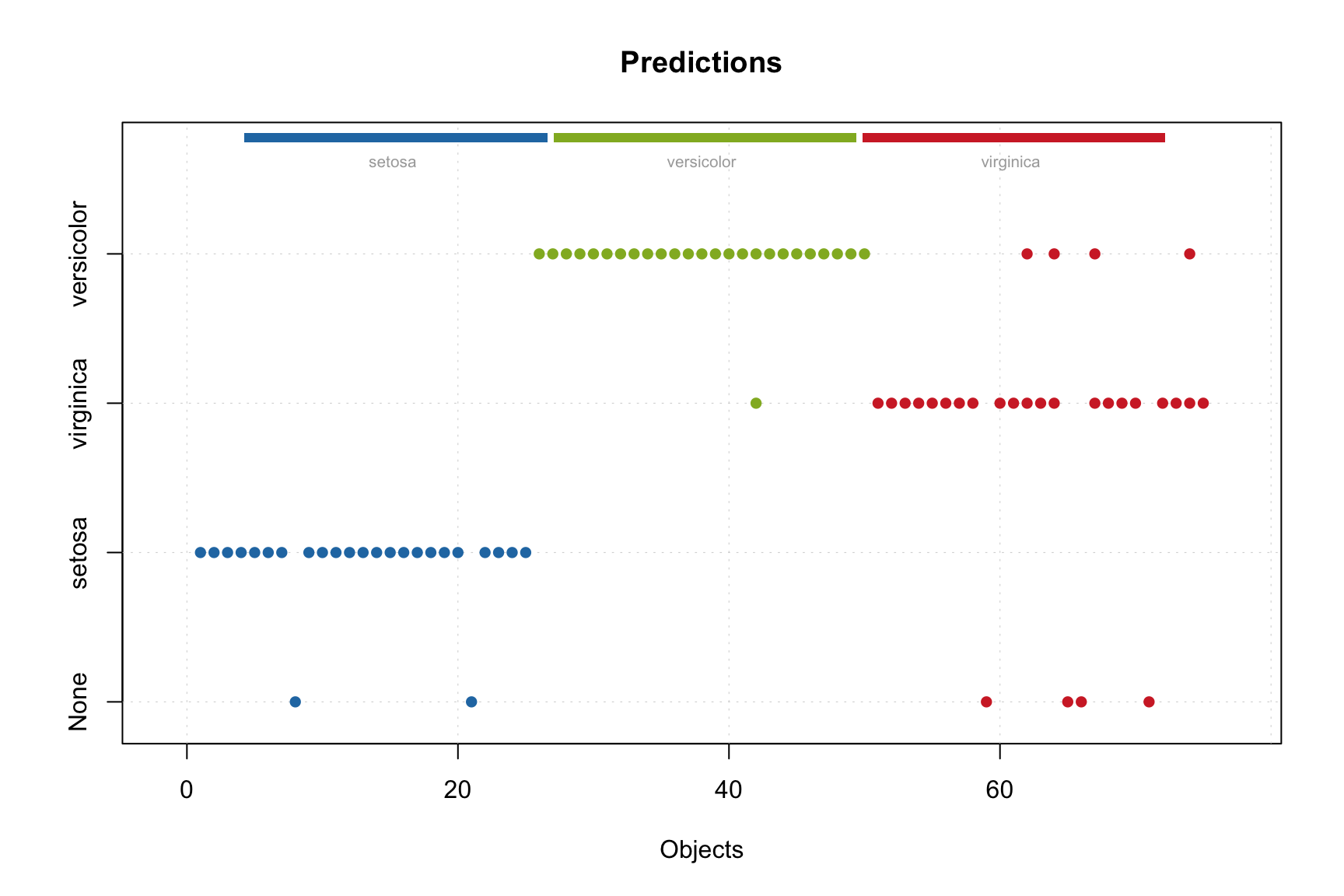
In this case, the predictions are shown only for the number of components each model found optimal. The names of classes along y-axis are the individual models. Similarly we can show the predicted values.
## setosa virginica versicolor
## 40 1 -1 -1
## 42 -1 -1 -1
## 44 1 -1 -1
## 46 1 -1 -1
## 48 1 -1 -1
## 50 1 -1 -1
## 52 -1 -1 1
## 54 -1 -1 1
## 56 -1 -1 1
## 58 -1 -1 1
## 60 -1 -1 1Method getConfusionMatrix() is also available in this case.
## setosa virginica versicolor None
## setosa 23 0 0 2
## virginica 0 21 4 4
## versicolor 0 1 25 0There are three additional plots available for multiclass SIMCA model. First of all it is a distance between a selected model and the others.
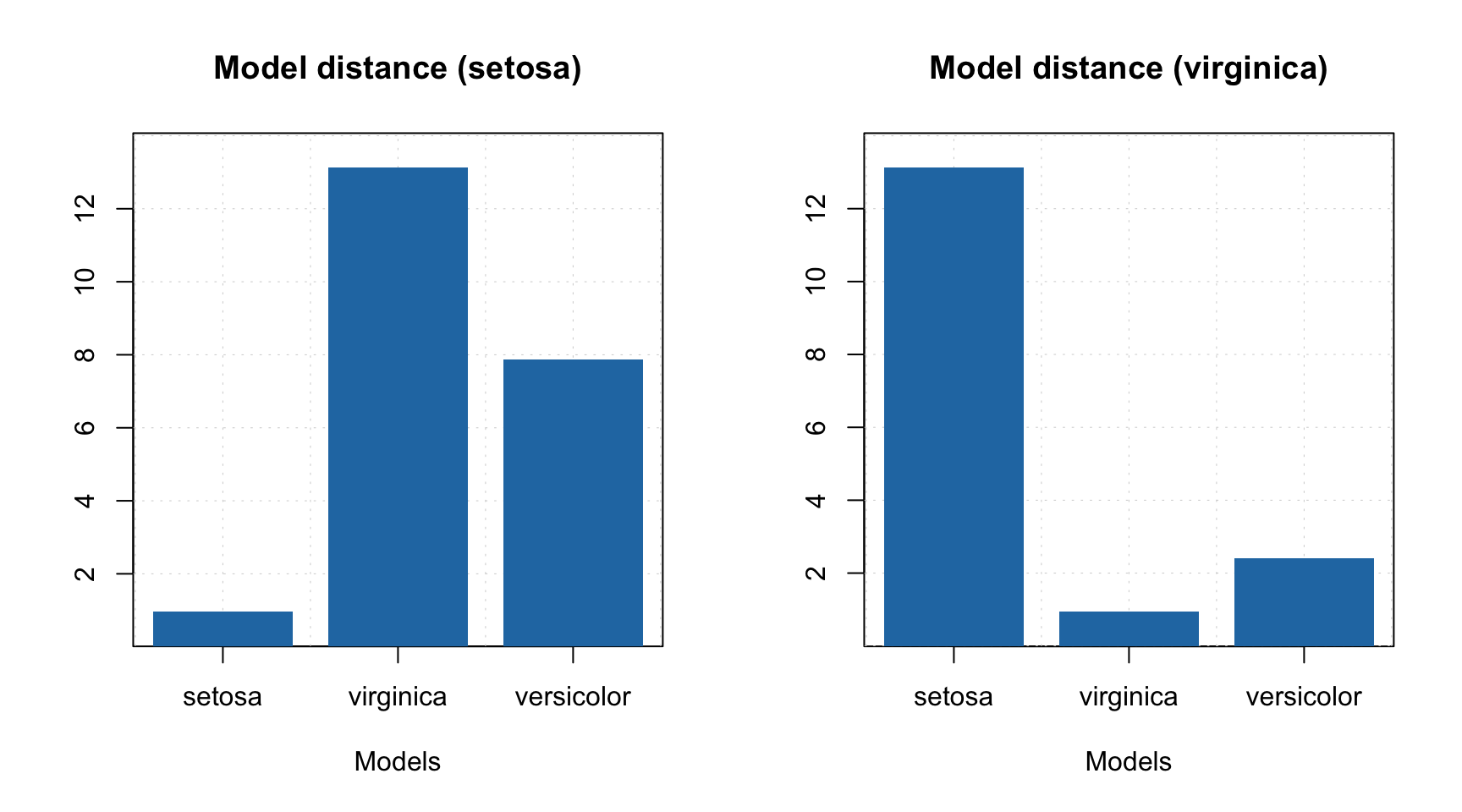 The plot shows not a real distance but rather a similarity between a selected model and the others as a ratio of residual variances. You can find more detailed description about how the model distance is calculated in description of the method or in help for
The plot shows not a real distance but rather a similarity between a selected model and the others as a ratio of residual variances. You can find more detailed description about how the model distance is calculated in description of the method or in help for plotModelDistance.simcam function.
The second plot is a discrimination power, mentioned in the beginning of the section.
par(mfrow = c(1, 2))
plotDiscriminationPower(mm, c(1, 3), show.labels = TRUE)
plotDiscriminationPower(mm, c(2, 3), show.labels = TRUE)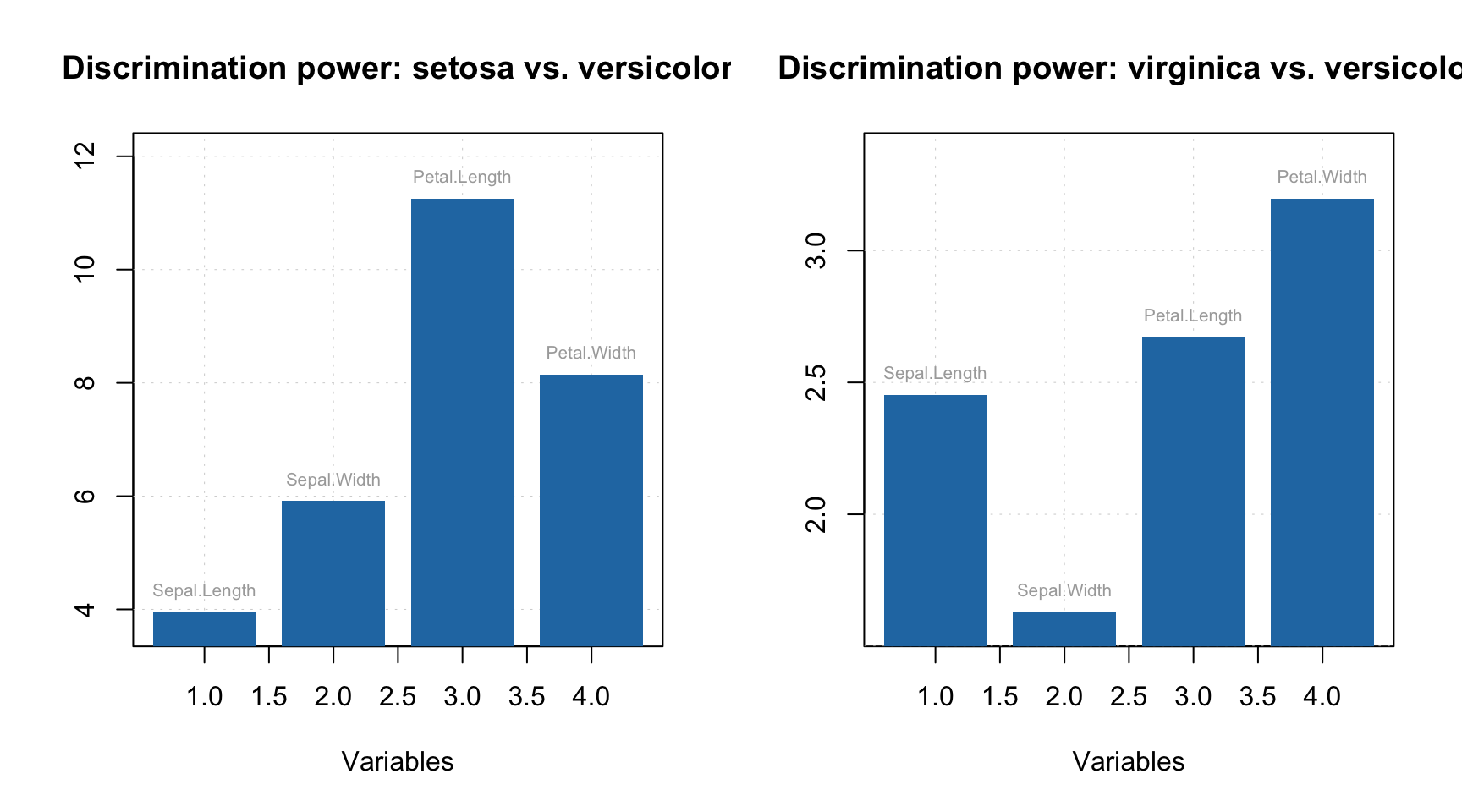
And, finally, a Cooman’s plot showing an orthogonal distance, \(q\), from objects to two selected classes/models.
par(mfrow = c(1, 2))
plotCooman(mm, c(1, 3), show.labels = TRUE)
plotCooman(mm, c(2, 3), show.labels = TRUE)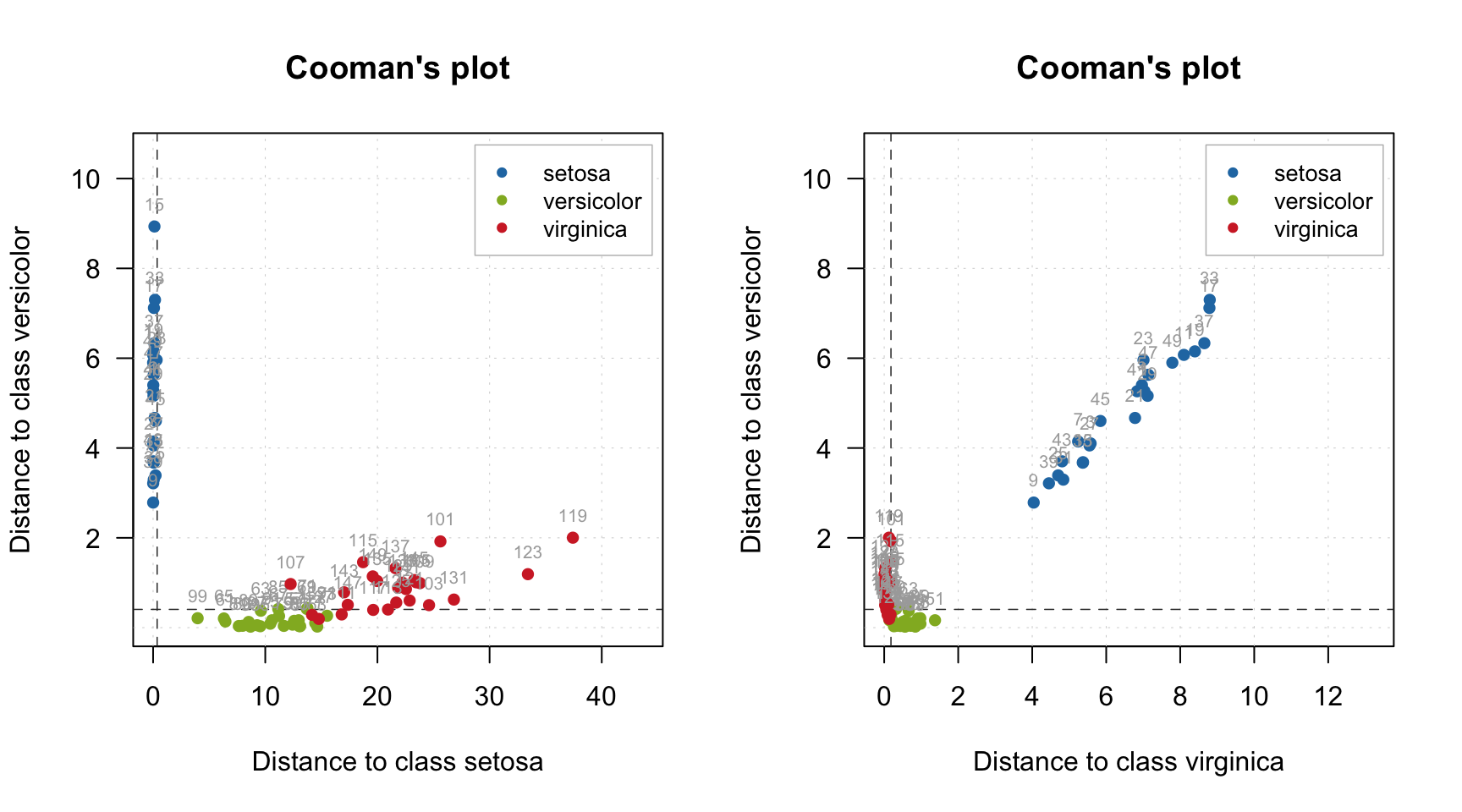
The limits, shown as dashed lines, are computed using chi-square distribution but only for \(q\) values.