Correction of spectral baseline
Baseline correction methods include Extended Multiplicative Scatter Correction (EMSC) and correction of baseline with Asymmetric Least Squares (ALS). Another common method for baseline correction, Standard Normal Variate (SNV), is a part of normalization methods described in the next section.
Extended Multiplicative Scatter Correction
Multiplicative Scatter Correction is a simple procedure aiming first of all at removing additive and multiplicative scatter effects from Vis/NIR spectra. The Extended MSC does this plus corrects non-linear effects of the baseline. The theoretical foundation of the method can be found in this paper. In mdatools the method is implemented as function prep.emsc().
The main parameter of the function is degree that defines the polynomial degree used to fit the baseline. If degree=0, the EMSC is simplified to the conventional MSC method. Both MSC and EMSC rely on a reference spectrum. Usually a mean spectrum of a training set is used for this purpose. If you do not provide the reference spectrum manually (via parameter mspectrum), it will be calculated automatically for all rows of the provided data matrix.
Here are some examples:
# set random generator to get reproducible results
set.seed(42)
# load UV/Vis spectra from Simdata
data(simdata)
# add attributes to make plots more informative
w = simdata$wavelength
spectra0 = simdata$spectra.c
attr(spectra0, "xaxis.values") = w
attr(spectra0, "xaxis.name") = "Wavelength, nm"
# save number of spectra (rows) and number of wavelength (columns)
n = nrow(spectra0)
m = ncol(spectra0)
# add random additive and multiplicative effects
A = matrix(rnorm(n, 0, 0.0005), n, m) # multiplicative effect
B = matrix(rnorm(n, 0, 0.01), n, m) # additive effect
X = matrix(seq_len(m), n, m, byrow = TRUE) # the baseline
spectra1 = spectra0 + A * X + B
# add attributes to the distorted spectra
attr(spectra1, "xaxis.values") = w
attr(spectra1, "xaxis.name") = "Wavelength, nm"
# correct the spectra using MSC and EMSC with quadratic polynomial
spectra2 = prep.emsc(spectra1)
spectra3 = prep.emsc(spectra1, degree = 2)
# show the original, distorted and the preprocessed spectra
par(mfrow = c(2, 2))
mdaplot(spectra0, type = "l", main = "Original spectra")
mdaplot(spectra1, type = "l", main = "Distorted spectra")
mdaplot(spectra2, type = "l", main = "EMSC preprocessed (degree = 0)")
mdaplot(spectra3, type = "l", main = "EMSC preprocessed (degree = 2)")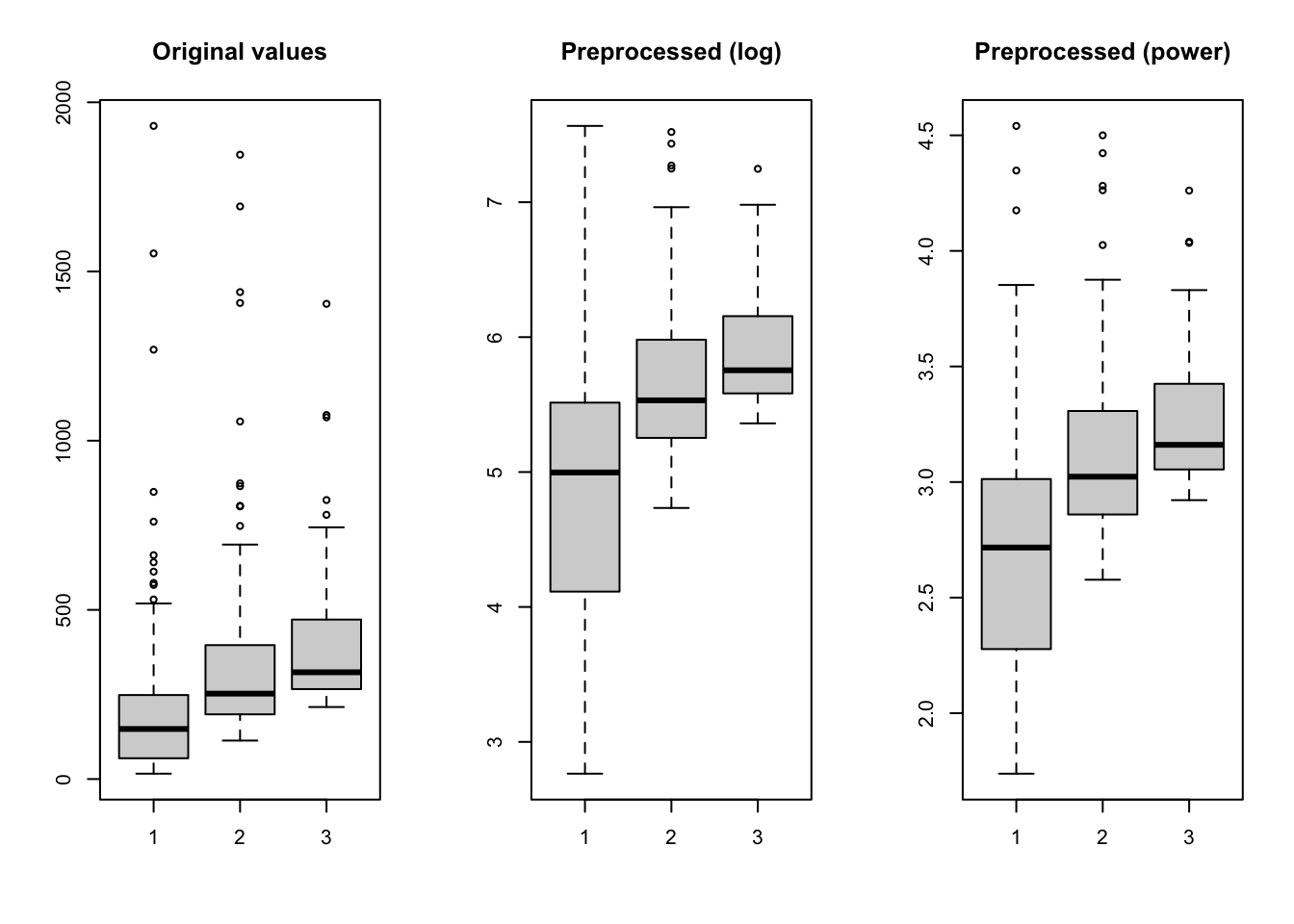
Asymmetric least squares
Asymmetric least squares (ALS) baseline correction allows correcting baseline issues, which have a wider shape compared to the characteristic peaks. It can be used for example to correct the fluorescence effect in Raman spectra.
The method is based on Whittaker smoother and was proposed in this paper. It is implemented as a function prep.alsbasecorr(), which has two main parameters — power of a penalty parameter
(plambda, usually varies between 2 and 9) and the ratio of asymmetry (p, usually between 0.1 and 0.001). For example, if plambda = 5, the penalty parameter \(\lambda\), described in the paper will be equal to \(10^5\).
The choice of the parameters depends on how broad the disturbances of the baseline are and how narrow the original spectral peaks are. In the example below we took original spectra from the carbs dataset, added baseline disturbance using broad Gaussian peaks and then tried to remove the disturbance by applying the prep.alsbasecorr(). The result is shown in the form of plots.
library(mdatools)
data(carbs)
# take original spectra from carbs dataset
x <- t(carbs$S)
# add disturbance to the baseline by using broad Gaussian peaks
y <- x + rbind(
dnorm(1:ncol(x), 750, 200) * 10000,
dnorm(1:ncol(x), 750, 100) * 10000,
dnorm(1:ncol(x), 500, 100) * 10000
)
# preprocess the disturbed spectra using ALS baseline correction
y.new <- prep.alsbasecorr(y, plambda = 5, p = 0.01)
# show the original, disturbed and the preprocessed spectra separately for each component
par(mfrow = c(3, 1))
for (i in 1:3) {
mdaplotg(list(
original = x[i, , drop = FALSE],
disturbed = y[i, , drop = FALSE],
preprocessed = y.new[i, , drop = FALSE]
), type = "l", lty = c(2, 1, 1), col = c("black", "red", "blue"),
main = paste("Pure component #", i)
)
}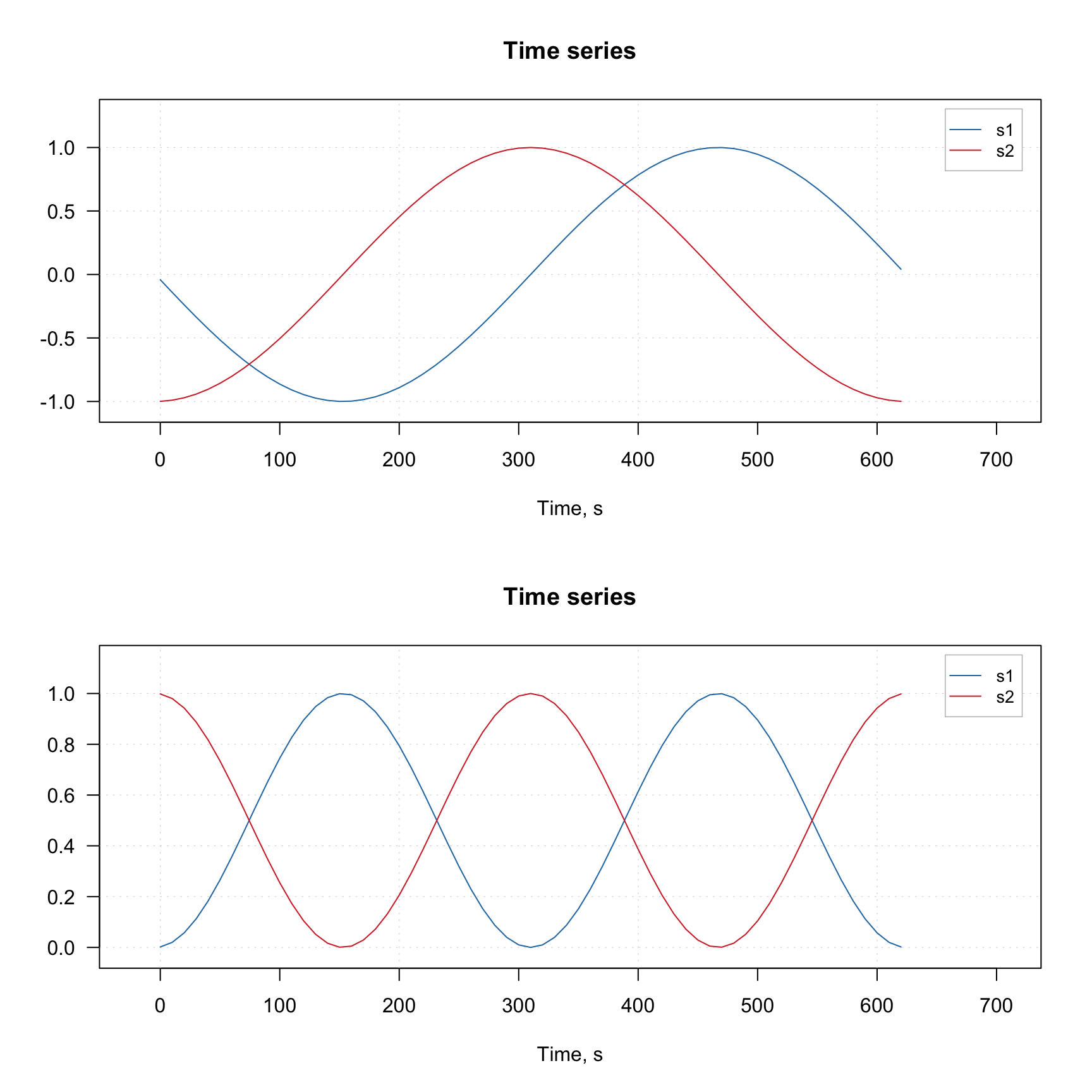
As one can notice, the blue curves with corrected spectra are pretty similar to the original spectra shown as dashed black curves.